Article information
Author: Anon until publication
This article is an unpublished pre-print undergoing public peer review organised by the WikiJournal of Science.
You can follow its progress through the peer review process at this tracking page.First submitted:
Reviewer comments
Author info:
QID: Q100272642
Suggested (provisional) citation format:
"Structural Model of Bacteriophage T4", WikiJournal preprints, Wikidata Q100272642
License: ![]()
![]() This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction, provided the original author and source are credited.
This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction, provided the original author and source are credited.
Thomas Shafee
Gorla Praveen contact Reviewers: (comments)
anonymous
Abstract
Introduction
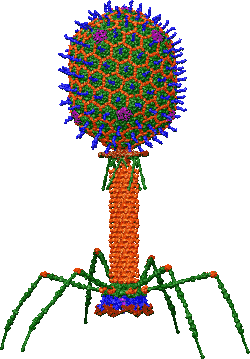
The complete structural model for bacteriophage T4 has been possible thanks to the determination of the structures of single proteins that constitute the virus as well as various parts of the virus. First, the capsid (head) of the virus was constructed using a 3D cryoEM reconstruction of the head where each single protein was fitted in the cryoEM (soc (orange), hoc (blue), gp23 (green), gp24 (magenta) and gp20 (red, hidden because is between the head and tail)). Then, similar procedure followed for the tail and tail fibers.[1][2] Every protein has been described in detail; even though this structural model is not perfect, it is the best reconstruction of the entire organism available in 2020.
Reconstruction
The reconstruction was possible using the molecular visualization software UCSF Chimera[3] in a computer and for some tasks in supercomputers like Bridges for Pittsburgh and Frontera from Texas. This structural model has been constructed during many years therefore is the accumulated work of many years of research in Catholic University of America.
There are approximately 50 structural proteins that assemble the virus constructed with protein databank (pdb) structures and one cryoEM reconstruction corresponding to the brown ring between head and tail. This structure can be used for teaching at any level of education and for research as well because is accurate at atomic resolution and therefore many suggestions and studies can be derived from it (e.g. how antigen-display technology could be used with bacteriophage t4).
Additional information
Acknowledgements
- This work used the Extreme Science and Engineering Discovery Environment (XSEDE),[4] which is supported by National Science Foundation grant number ACI-1548562. Specifically, it used the Bridges system,[5] which is supported by NSF award number ACI-1445606, at the Pittsburgh Supercomputing Center (PSC).
- Stanzione, Dan; West, John; Evans, R. Todd; Minyard, Tommy; Ghattas, Omar; Panda, Dhabaleswar K. (2020-07-26). "Frontera: The Evolution of Leadership Computing at the National Science Foundation". Practice and Experience in Advanced Research Computing (Portland OR USA: ACM): 106–111. doi:10.1145/3311790.3396656. ISBN 978-1-4503-6689-2. https://dl.acm.org/doi/10.1145/3311790.3396656.
Competing interests
The author declares no competing interests.
References
- ↑ Mesyanzhinov, V. V.; Leiman, P. G.; Kostyuchenko, V. A.; Kurochkina, L. P.; Miroshnikov, K. A.; Sykilinda, N. N.; Shneider, M. M. (2004-11). "Molecular architecture of bacteriophage T4". Biochemistry (Moscow) 69 (11): 1190–1202. doi:10.1007/s10541-005-0064-9. ISSN 0006-2979. http://link.springer.com/10.1007/s10541-005-0064-9.
- ↑ Taylor, Nicholas M. I.; Prokhorov, Nikolai S.; Guerrero-Ferreira, Ricardo C.; Shneider, Mikhail M.; Browning, Christopher; Goldie, Kenneth N.; Stahlberg, Henning; Leiman, Petr G. (2016-05). "Structure of the T4 baseplate and its function in triggering sheath contraction". Nature 533 (7603): 346–352. doi:10.1038/nature17971. ISSN 0028-0836. http://www.nature.com/articles/nature17971.
- ↑ Pettersen, Eric F.; Goddard, Thomas D.; Huang, Conrad C.; Couch, Gregory S.; Greenblatt, Daniel M.; Meng, Elaine C.; Ferrin, Thomas E. (2004-10). "UCSF Chimera—A visualization system for exploratory research and analysis". Journal of Computational Chemistry 25 (13): 1605–1612. doi:10.1002/jcc.20084. ISSN 0192-8651. http://doi.wiley.com/10.1002/jcc.20084.
- ↑ Towns, John; Cockerill, Timothy; Dahan, Maytal; Foster, Ian; Gaither, Kelly; Grimshaw, Andrew; Hazlewood, Victor; Lathrop, Scott et al. (2014-09). "XSEDE: Accelerating Scientific Discovery". Computing in Science & Engineering 16 (5): 62–74. doi:10.1109/MCSE.2014.80. ISSN 1521-9615. https://ieeexplore.ieee.org/document/6866038/.
- ↑ Nystrom, Nicholas A.; Levine, Michael J.; Roskies, Ralph Z.; Scott, J. Ray (2015). "Bridges: a uniquely flexible HPC resource for new communities and data analytics". Proceedings of the 2015 XSEDE Conference on Scientific Advancements Enabled by Enhanced Cyberinfrastructure - XSEDE '15. St. Louis, Missouri: ACM Press. pp. 1–8. doi:10.1145/2792745.2792775. ISBN 978-1-4503-3720-5.
