In computational geometry, a well-separated pair decomposition (WSPD) of a set of points , is a sequence of pairs of sets , such that each pair is well-separated, and for each two distinct points , there exists precisely one pair which separates the two.
The graph induced by a well-separated pair decomposition can serve as a k-spanner of the complete Euclidean graph, and is useful in approximating solutions to several problems pertaining to this.[1]
Definition
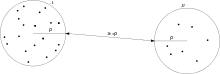
Let be two disjoint sets of points in , denote the axis-aligned minimum bounding box for the points in , and denote the separation factor.
We consider and to be well-separated, if for each of and there exists a d-ball of radius containing it, such that the two spheres have a minimum distance of at least .[2]
We consider a sequence of well-separated pairs of subsets of , to be a well-separated pair decomposition (WSPD) of if for any two distinct points , there exists precisely one , , such that either
- and , or
- and .[1]
Construction
Split tree
By way of constructing a fair split tree, it is possible to construct a WSPD of size in time.[2]
The general principle of the split tree of a point set S is that each node u of the tree represents a set of points Su and that the bounding box R(Su) of Su is split along its longest side in two equal parts which form the two children of u and their point set. It is done recursively until there is only one point in the set.
Let Lmax(R(X)) denote the size of the longest interval of the bounding hyperrectangle of point set X and let Li(R(X)) denote the size of the i-th dimension of the bounding hyperrectangle of point set X. We give pseudocode for the Split tree computation below.
SplitTree(S) Let u be the node for S if |S| = 1 R(u) := R(S) // R(S) is a hyperrectangle which each side has a length of zero. Store in u the only point in S. else Compute R(S) Let the i-th dimension be the one where Lmax(R(S)) = Li(R(S)) Split R(S) along the i-th dimension in two same-size hyperrectangles and take the points contained in these hyperrectangles to form the two sets Sv and Sw. v := SplitTree(Sv) w := SplitTree(Sw) Store v and w as, respectively, the left and right children of u. R(u) := R(S) return u
This algorithm runs in time.
We give a more efficient algorithm that runs in time below. The goal is to loop over the list in only operations per step of the recursion but only call the recursion on at most half the points each time.
Let Sij be the i-th coordinate of the j-th point in S such that Si is sorted according to the i-th coordinate and p(Sij) be the point. Also, let h(R(S)) be the hyperplane that splits the longest side of R(S) in two. Here is the algorithm in pseudo-code:
SplitTree(S, u) if |S| = 1 R(u) := R(S) // R(S) is a hyperrectangle which each side has a length of zero. Store in u the only point in S. else size := |S| repeat Compute R(S) R(u) := R(S) j : = 1 k : = |S| Let the i-th dimension be the one where Lmax(R(S)) = Li(R(S)) Sv : = ∅ Sw : = ∅ while Sij+1 < h(R(S)) and Sik-1 > h(R(S)) size := size - 1 Sv : = Sv ∪ {p(S_i^j)} Sw : = Sw ∪ {p(S_i^k)} j := j + 1 k := k - 1 Let v and w be respectively, the left and right children of u. if Sij+1 > h(R(S)) Sw := S \ Sv u := w S := Sw SplitTree(Sv,v) else if Sik-1 < h(R(S)) Sv := S \ Sw u := v S := Sv SplitTree(Sw,w) until size ≤ n⁄2 SplitTree(S,u)
To be able to maintain the sorted lists for each node, linked lists are used. Cross-pointers are kept for each list to the others to be able to retrieve a point in constant time. In the algorithm above, in each iteration of the loop, a call to the recursion is done. In reality, to be able to reconstruct the list without the overhead of resorting the points, it is necessary to rebuild the sorted lists once all points have been assigned to their nodes. To do the rebuilding, walk along each list for each dimension, add each point to the corresponding list of its nodes, and add cross-pointers in the original list to be able to add the cross-pointers for the new lists. Finally, call the recursion on each node and his set.
WSPD computation

The WSPD can be extracted from such a split tree by calling the recursive FindPairs(v,w) function on the children of every node in the split tree. Let ul / ur denote the children of the node u. We give pseudocode for the FindWSPD(T, s) function below.
FindWSPD(T, s)
for each node u that is not a leaf in the split tree T do
FindPairs(ul, ur)
We give pseudocode for the FindPairs(v, w) function below.
FindPairs(v, w)
if Sv and Sw are well-separated with respect to s
report pair(Sv, Sw)
else
if( Lmax(R(v)) ≤ Lmax(R(w)) )
Recursively call FindPairs(v, wl) and FindPairs(v, wr)
else
Recursively call FindPairs(vl, w) and FindPairs(vr, w)
Combining the s-well-separated pairs from all the calls of FindPairs(v,w) gives the WSPD for separation s.
Proof of correctness of the algorithm |
|---|
|
It is clear that the pairs returned by the algorithm are well-separated because of the return condition of the function FindPairs. Now, we have to prove that for any distinct points and in , there is a unique pair so that (i) and or (ii) and . Assume without loss of generality that (i) holds. Let be the lowest common ancestor of and in the split tree and let and be the children of . Because of the last assumption, is in the subtree of and in the subtree of . A call to FindPairs(v,w) is necessarily done in FindWSPD. Because, each time there is a recursion, the recursion tree creates two branches that contain all the points of the current recursion call, there will be a sequence of call to FindPairs leading to having in and in . Because is the lowest common ancestor of and , calling FindPairs on the children of a higher node would result of and not being in a pair and calling FindPairs on the children in one of the nodes of one of the subtrees of would result by or not being in any pair. Thus, the pair is the unique one separating and . |
Each time the recursion tree split in two, there is one more pair added to the decomposition. So, the algorithm run-time is in the number of pairs in the final decomposition.
Callahan and Kosaraju proved that this algorithm finds a Well-separated pair decomposition (WSPD) of size .[2]
Properties
Lemma 1: Let be a well-separated pair with respect to . Let and . Then, .
Proof: Because and are in the same set, we have that where is the radius of the enclosing circle of and . Because and are in two well-separated sets, we have that . We obtain that:
Lemma 2: Let be a well-separated pair with respect to . Let and . Then, .
Proof: By the triangle inequality, we have:
From Lemma 1, we obtain:
Applications
The well-separated pair decomposition has application in solving a number of problems. WSPD can be used to:
- Solve the closest pair problem in time.[1]
- Solve the k-closest pairs problem in time.[1]
- Solve the k-closest pair problem in time.[3]
- Solve the all-nearest neighbors problem in time.[1]
- Provide a -approximation of the diameter of a point set in time.[1]
- Directly induce a t-spanner of a point set.[1]
- Provide a t-approximation of the Euclidean minimum spanning tree in d dimensions in time.[1]
- Provide a -approximation of the Euclidean minimum spanning tree in d dimensions in time.[4]
References
- 1 2 3 4 5 6 7 8 Smid, Michiel (16 August 2005). "The well-separated pair decomposition and its applications" (PDF). Retrieved 26 March 2014.
- 1 2 3 Callahan, P. B. & Kosaraju, S. R. (January 1995). "A Decomposition of Multidimensional Point Sets with Applications to k-Nearest-Neighbors and n-Body Potential Fields". Journal of the ACM. 42 (1): 67–90. doi:10.1145/200836.200853.
- ↑ Bespamyatnikh, Sergei; Segal, Michael (2002). "Fast Algorithms for Approximating Distances". Algorithmica. 33 (2): 263–269. doi:10.1007/s00453-001-0114-7.
- ↑ Arya, Sunil; Mount, David M. (2016). "A fast and simple algorithm for computing approximate euclidean minimum spanning trees". Proceedings of the Twenty-Seventh Annual ACM-SIAM Symposium on Discrete Algorithms.