Leukocyte Receptor Cluster Member 9 (LENG 9) is an uncharacterized protein encoded by the LENG9 gene.[1][2] In humans, LENG9 is predicted to play a role in fertility and reproductive disorders associated with female endometrium structures.[3][4]
Gene
Location

LENG9 is located at 19q13.42 on chromosome 19, spanning the sense strand (-) from 54,461,796 bp to 54,463,711 bp.[5] The LENG9 gene is 1,930 base pairs in length and contains one exon.[1][2]
Gene Neighborhood
Genes LENG8-AS1 and CDC42EP5 neighbor LENG9 on chromosome 19.[2] CDC42EP5 extends over the same region of LENG9 while LENG8-AS1 is located to the left of both genes.[5] TTYH1 and LENG8 are also found in the same gene neighborhood but are located on the opposite strand.
Expression

LENG9 is highly expressed (75-100%) in skeletal muscles and part of fetal liver tissues while ubiquitous expression of LENG9 is moderate (50-75%) in all other tissues observed.[6][7] Human expression of LENG9 is observed in the cervix, lung, and placenta of adults.[8] The gene is also expressed in disease states including lung tumors and primitive neuroectodermal tumors, usually found in children or young adults. However, LENG9 is not expressed during the juvenile stage of development.[8]
Promoter
The promoter region is predicted to be 1101 base pairs in length.[9] The transcriptional start site found in this region is located 119 bp upstream of the start codon[1] as well as an in-frame stop codon at 1087 bp to 1089 bp.[5]
mRNA Transcript
Splice Variants
In humans, LENG9 has two mRNA unspliced transcript variants.[5] Variant (1) is the longest and most conserved transcript of the gene and is made up of one exon that is composed of 1,919 bp.
Protein
General Properties
LENG9 is 501 amino acids in length, with a predicted molecular weight of 53.2 kDa.[10] The isoelectric point of LENG9 protein is predicted to be 7.7.[11] No known transmembrane sequences were found for LENG9.[12]
Composition
Analysis of the LENG9 protein was performed against the "human" database,[10] which indicated a higher frequency of alanine and proline amino acids than of that of a normal human protein. Inversely, an abnormally lower frequency of aspartate, isoleucine, methionine, asparagine, serine, and tyrosine amino acids were detected.
Structure
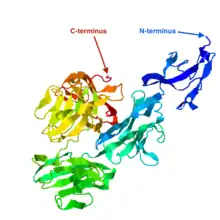
The secondary structure of LENG9 is predicted to be composed of alpha-helices and beta-sheets throughout the sequence.[14][13][15] The tertiary structure of LENG9 is displayed in the image to the right.
Sub-cellular localization
A strong signal peptide detected in the mitochondrion region (0.788) suggests that the LENG9 protein localizes in the mitochondrial matrix.[12] However, further analysis among other mammal orthologs predicted sub-cellular localization in the cytoplasm and nuclear localization for Danio rerio.[12][16]
Post Translational Modifications

LENG9 is predicted to undergo post-translational modifications such as phosphorylation, N-terminal acetylation, sumoylation, and C-terminal Glycosylphosphatidylinositol (GPI) anchor modification.
Phosphorylation
LENG9 contains numerous phosphorylation sites distributed in the protein sequence, as show in the diagram to the right. These sites include casein kinase 2 (CK2), cAMP-dependent protein kinase (PKA), protein kinase C (PKC), ataxia telangiectasia mutated kinase (ATM), cyclin-dependent kinase 5 (CDK5), and casein kinase 1 (CK1).[18]
N-terminal Acetylation
There is one predicted N-terminal acetylation site found in the protein LENG9 at the serine amino acid position 3.[19]
Sumoylation
There are two predicted sumoylation sites within LENG9 at position 82 and 452 on lysine residues.[20]
C-Terminal GPI Anchor Modification
A C-terminal GPI modification site was found on the glycine residue at position 486.[21]
Domains and Motifs
There are 3 conserved domains in LENG9. The metal-ion binding zinc finger domain ZnF_C3H1[22] is found from amino acid 46 to 61. LENG9 also has a domain of unknown function belonging to the domain family DUF504, that spans from amino acid 109 to 160. The last conserved domain spans from amino acid 320 to 500 and is known as AKAP7 2'5' RNA ligase-like domain (AKAP7_NLS).[5][23]
Conserved WD40 repeats are found in LENG9, spanning from amino acid 98 to 159.[13][24] This motif is characterized by beta-propeller structures in the tertiary structure.

Protein Interactions
LENG9 is found to interact with the C9ORF41 protein that encodes methyltransferase, involved in converting carnosine to anserine present in skeletal muscle. Another interactor is CDC5L,[26] which is a positive regulator for the cell cycle for phase G2 to M transition.[27] FOXS1 is another interacting protein that functions as a transcriptional repressor to suppress promoters such as FASLG, FOXO3, and FOXO4.[28]
Clinical Significance
Disease Association
The LENG9 gene was found to be up-regulated and expressed in endothelial endometrium (hEECs) tissues during various stages of the menstrual/reproductive cycle when transfected with the RNA gene, miR-30d.[3][29] As ectopic over-expression of miR-30d in hEECs is observed to affect cancer-associated genes, LENG9 is predicted to play a role in reproductive and endocrine system disorders.
Fertility
Analysis of endometrial receptivity using miRNA of receptive and prereceptive endometrium from fertile women also indicated significant up-regulation of miR-30d.[4] Consequently, the induced expression of LENG9 from miR-30d transfection suggests a possible relationship between the LENG9 gene and female fertility functions.
Homology

Evolution
Comparison of the LENG9 protein was conducted against fibrinogen and cytochrome C to observe the rate of evolutionary change. The relatively fast rate of change in LENG9 compared to that of other proteins suggests that the gene is adaptive for vital cell structures and functions.
Paralogs
There are no known paralogs for LENG9.[2]
Orthologs
LENG9 is highly conserved in mammals and bony fish such as the zebrafish.[24] It is also conserved a in few reptiles and amphibians.[31] The gene is not present in invertebrates, birds, bacteria, or fungi[32]
| Genus and Species | Common Name | Order | Divergence
from Human Lineage (MYA) |
NCBI Accession
Number |
Sequence Length
(bp) |
Percent Identity
to Human |
Percent Similarity to Human |
|---|---|---|---|---|---|---|---|
| Homo sapiens | Human | Primates | 0 | NP_945339.2 | 501 | 100% | 100% |
| Pan troglodytes | Chimpanzee | 6.65 | XP_003316725.1 | 479 | 94.4% | 94.4% | |
| Gorilla gorilla gorilla | Gorilla | 9.06 | XP_018870227.1 | 458 | 89.4% | 89.8% | |
| Macaca mulatta | Rhesus Macaque | 29.44 | XP_014980381.1 | 476 | 80.7% | 83.5% | |
| Ictidomys tridecemlineatus | Thirteen-Lined Squirrel | Rodentia | 90 | XP_005341645.1 | 490 | 62.1% | 68.2% |
| Peromyscus maniculatus bairdii | Deer Mouse | 90 | XP_006995933.1 | 488 | 56.6% | 63.5% | |
| Ceratotherium simum simum | Southern White Rhinoceros | Perissodactyla | 96 | XP_014649760.1 | 528 | 58.3% | 64.1% |
| Equus caballus | Horse | 96 | XP_001488865.2 | 506 | 58.0% | 63.5% | |
| Odobenus rosmarus divergens | Walrus | Carnivora | 94 | XP_004415925.1 | 527 | 59.3% | 62.5% |
| Panthera pardus | Leopard | 96 | XP_019292295.1 | 540 | 55.1% | 61.0% | |
| Bos taurus | Cattle | Artiodactyla | 96 | XP_005192875.1 | 535 | 53.6% | 58.7% |
| Ovis aries | Sheep | 96 | XP_014955819.1 | 669 | 41.1% | 47.1% | |
| Alligator mississippiensis | American Alligator | Crocodilla | 312 | XP_014459626.1 | 452 | 39.0% | 46.5% |
| Thamnophis sirtalis | Common Garter Snake | Squamata | 312 | XP_013921249.1 | 505 | 32.6% | 45.5% |
| Xenopus laevis | African Clawed Frog | Anura | 352 | XP_018080040.1 | 431 | 30.9% | 40.6% |
| Nanorana parkeri | High Himalaya Frog | 352 | XP_018430267.1 | 401 | 29.4% | 39.3% | |
| Lates calcarifer | Barramundi | Perciformes | 435 | XP_018538114.1 | 565 | 31.3% | 38.2% |
| Nothobranchius furzeri | Tortoise Killfish | Cyprinodontiformes | 435 | XP_015805834.1 | 537 | 30.3% | 39.4% |
| Danio Rerio | Zebrafish | 435 | XP_005157957.1 | 542 | 27.9% | 37.5% | |
| Poecilia formosa | Amazon Molly | 435 | XP_007550061.1 | 569 | 26.7% | 37.5% |
References
- 1 2 3 4 "LENG9 leukocyte receptor cluster member 9 [Homo sapiens (human)] - Gene - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2017-05-06.
- 1 2 3 4 Database, GeneCards Human Gene. "LENG9 Gene - GeneCards | LENG9 Protein | LENG9 Antibody". www.genecards.org. Retrieved 2017-04-30.
- 1 2 Moreno-Moya, Juan Manuel; Vilella, Felipe; Martínez, Sebastián; Pellicer, Antonio; Simón, Carlos (2014-06-01). "The transcriptomic and proteomic effects of ectopic overexpression of miR-30d in human endometrial epithelial cells". MHR: Basic Science of Reproductive Medicine. 20 (6): 550–566. doi:10.1093/molehr/gau010. ISSN 1360-9947. PMID 24489115.
- 1 2 Altmäe, Signe; Martinez-Conejero, Jose A.; Esteban, Francisco J.; Ruiz-Alonso, Maria; Stavreus-Evers, Anneli; Horcajadas, Jose A.; Salumets, Andres (2012-08-17). "MicroRNAs miR-30b, miR-30d, and miR-494 Regulate Human Endometrial Receptivity". Reproductive Sciences. 20 (3): 308–317. doi:10.1177/1933719112453507. PMC 4077381. PMID 22902743.
- 1 2 3 4 5 "LENG9 leukocyte receptor cluster member 9 [Homo sapiens (human)] - Gene - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2017-02-19.
- 1 2 "49016057 - GEO Profiles - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2017-05-06.
- ↑ "Leukocyte receptor cluster (LRC) member 9 (LENG9)". www.ncbi.nlm.nih.gov. Retrieved 2017-05-06.
- 1 2 Group, Schuler. "EST Profile - Hs.590976". www.ncbi.nlm.nih.gov. Retrieved 2017-05-06.
- ↑ "Genomatix: Annotation & Analysis". www.genomatix.de. Retrieved 2017-04-30.
- 1 2 "SAPS". Biology Workbench. Retrieved May 6, 2017.
- ↑ "PI". Retrieved May 6, 2017.
- 1 2 3 "TargetP 1.1 Server". www.cbs.dtu.dk. Retrieved 2017-04-30.
- 1 2 3 "I-TASSER results". zhanglab.ccmb.med.umich.edu. Retrieved 2017-05-06.
- ↑ "Phyre 2 Results for Undefined". www.sbg.bio.ic.ac.uk. Retrieved 2017-04-30.
- ↑ "CHOFAS". Biology Workbench. Retrieved May 6, 2017.
- ↑ "PSORT II Prediction". psort.hgc.jp. Retrieved 2017-05-06.
- ↑ "NetPhos 3.1 Server". www.cbs.dtu.dk. Retrieved 2017-05-07.
- ↑ "NetPhos 3.1 Server". www.cbs.dtu.dk. Retrieved 2017-05-06.
- ↑ "NetAcet 1.0 Server". www.cbs.dtu.dk. Retrieved 2017-05-06.
- ↑ "SUMOplot™ Analysis Program | Abgent".
- ↑ "GPI Prediction Server". mendel.imp.ac.at. Retrieved 2017-05-06.
- ↑ "SMART: ZnF_C3H1 domain annotation". smart.embl.de. Retrieved 2017-05-06.
- ↑ "leukocyte receptor cluster member 9 isoform 1 [Homo sapiens] - Protein - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2017-05-06.
- 1 2 "CLUSTALW". Biology Workbench. Retrieved May 6, 2017.
- ↑ Castro, Edouard de. "PROSITE". prosite.expasy.org. Retrieved 2017-05-06.
- ↑ Huttlin, Edward L.; Ting, Lily; Bruckner, Raphael J.; Gebreab, Fana; Gygi, Melanie P.; Szpyt, John; Tam, Stanley; Zarraga, Gabriela; Colby, Greg (2015-07-16). "The BioPlex Network: A Systematic Exploration of the Human Interactome". Cell. 162 (2): 425–440. doi:10.1016/j.cell.2015.06.043. ISSN 1097-4172. PMC 4617211. PMID 26186194.
- ↑ Llères, David; Denegri, Marco; Biggiogera, Marco; Ajuh, Paul; Lamond, Angus I. (2010-06-01). "Direct interaction between hnRNP-M and CDC5L/PLRG1 proteins affects alternative splice site choice". EMBO Reports. 11 (6): 445–451. doi:10.1038/embor.2010.64. ISSN 1469-3178. PMC 2892320. PMID 20467437.
- ↑ Li, Xu; Wang, Wenqi; Wang, Jiadong; Malovannaya, Anna; Xi, Yuanxin; Li, Wei; Guerra, Rudy; Hawke, David H.; Qin, Jun (2015-01-21). "Proteomic analyses reveal distinct chromatin-associated and soluble transcription factor complexes". Molecular Systems Biology. 11 (1): 775. doi:10.15252/msb.20145504. ISSN 1744-4292. PMC 4332150. PMID 25609649.
- ↑ Database, GeneCards Human Gene. "MIR30D Gene - GeneCards | MIR30D RNA Gene". www.genecards.org. Retrieved 2017-05-07.
- ↑ "Protein BLAST: search protein databases using a protein query". blast.ncbi.nlm.nih.gov. Retrieved 2017-05-07.
- ↑ "Human BLAT Search". genome.ucsc.edu. Retrieved 2017-05-07.
- ↑ "Protein BLAST: search protein databases using a protein query". blast.ncbi.nlm.nih.gov. Retrieved 2017-05-06.