Hydrocarbonoclastic bacteria (also known as hydrocarbon degrading bacteria, oil degrading bacteria or HCB) are a heterogeneous group of prokaryotes which can degrade and utilize hydrocarbon compounds as source of carbon and energy. Despite being present in most of environments around the world, several of these specialized bacteria live in the sea and have been isolated from polluted seawater.
Taxonomy and distribution

The taxonomic diversity of hydrocarbon-degrading bacteria has not changed dramatically if we consider the higher taxa, many studies have provided information on 25 kinds of hydrocarbon-degrading bacteria and 25 kinds of fungi isolated from marine environments.[1] Bacterial genera such as Gordonia, Brevibacterium, Aeromicrobium, Dietzia, Burkholderia and Mycobacterium isolated from oil have been shown to be potential organisms for hydrocarbon degradation. Cerniglia et al. observed that nine cyanobacteria, five green algae, one red alga, one brown alga and two diatoms could oxidise naphthalene. Temperature is crucial because it influences microbial physiology and diversity; the rate of biodegradation generally decreases as the temperature decreases.[1]
Hydrocarbonoclastic bacteria are diazophilic, i.e. they can live in environments extremely poor in nitrogen compounds, which allows them to distribute themselves throughout the environment. They are extremely useful for environmentally friendly biosanitation; the fastest and most complete degradation occurs under aerobic conditions.
Hydrocarbons occur in marine environments where there are oil spills, which makes us understand that they are nutritionally independent of nitrogen sources, a characteristic due to their ability to fix atmospheric nitrogen. In Lagos in a city in Nigeria, nine bacterial strains Pseudomonas fluorescens, P. aeruginosa, Bacillus subtilis, Bacillus sp., Alcaligenes sp., Acinetobacter lwoffi, Flavobacterium sp., Micrococcus roseus and Corynebacterium sp, isolated from the polluted flow that could degrade crude oil, were detected ; in north-east India they were also detected. In the Louisiana incident in the Gulf of Mexico, about 100 strains were detected and studied, revealing that the isolates all belong to the phylum Proteobacteria and three classes (Alteromonadales, Rhodospirillales and Enterobacteriales).[2]
These organisms are normally present in very small numbers, which gives them an advantage over hydrocarbons such as carbon and energy, as they grow and multiply rapidly. Alcanivorax-like bacteria have been detected in oil-affected environments around the world, including the US, Germany, the UK, Spain, Italy, Singapore, China, the western Philippines, Japan, the mid-Atlantic ridge near Antarctica, and from deepwater sediments in the eastern Pacific Ocean.[3]
Physiology and metabolism
Hydrocarbonoclastic bacteria have two fundamental characteristics: (1) specific membrane-bound dioxygenases[4] and (2) mechanisms for optimizing contact with water-insoluble hydrocarbons.[5] Microbial biodegradation occurs wherever oil contamination occurs. However, biodegradation rates are slow and as a result there are severe toxic effects on marine life in the water and on the coast.[6]
The hydrocarbons contained in petroleum have a different behavior in water depending on their chemical nature. This process is called weathering, those with low molecular weight volatilize when they reach the surface.
The rest is attacked by bacteria that are able to do this. These bacteria do not adhere to the oil and do not have a high hydrophobicity of the cell surface. The next stage of degradation involves microorganisms with high cell surface hydrophobicity, which can adhere to residual high molecular weight hydrocarbons. Adhesion is due to hydrophobic fimbriae, fibrils, lipids and proteins of the outer membrane and some small molecules of the cell surface, such as gramicidin S and prodigiosin.[7]
All petroleum products are derived from crude oil whose major constituents are hydrocarbons, that can be separated into four fractions: saturated, aromatic, resin and asphaltene fractions. The susceptibility of hydrocarbons to microbial degradation can be generally ranked as follows: linear alkanes branched alkanes > small aromatics > cyclic alkanes.[8][9] Some compounds, such as the high molecular weight polycyclic aromatic hydrocarbons (PAHs), may not be degraded at all, asphaltenes and resins are considered to be recalcitrant to biodegradation.[10]
Alkane degradation pathways
Alkanes are readily biodegraded aerobically in the sea by several different pathways.
Terminal oxidation
The degradation of medium-length ones by Pseudomonas putida starts from the alkane hydroxylase, this enzyme is made up of three components: the membrane-bound oxygenase component and two soluble components called rubredoxin and rubredoxin reductase.[11]
From the oxidation of the methyl group of n-alkanes by the alkane hydroxylase, n-alkanols are released which are further oxidized by a membrane-bound alcohol dehydrogenase in n-alkanals. The n-alkanals are subsequently transformed into fatty acids and then into acyl CoA, respectively by the aldehyde dehydrogenase and by the acyl-CoA synthetase.
Subterminal oxidation
This path leads to the release of secondary alcohols. The n-alkanes are oxidized by monooxygenase to secondary alcohols, then to ketones and finally to fatty acids.[12]
Cycloalkane degradation pathways
Cycloalkanes are degraded by a co-oxidation mechanism, the process leading to the formation of a cyclic alcohol and a ketone. A monooxygenase introduces an oxygen into the cyclic ketone and the cyclic ring is cleaved.[13]
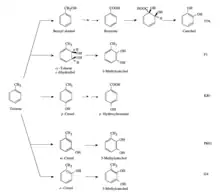
Aromatic hydrocarbon degradation pathways
For aromatic compounds there are different pathways, considering toluene at least five are known,[14] each of these is present in specific bacterial species, Burkholderia sp. strain JS150 is unique in using multiple pathways for toluene metabolism:[15]
- TOL pathway: This path takes the name of the homonymous plasmid that codes for it. Toluene is degraded to alcohol to benzyl, to benzaldehyde and then to benzoate, which is further transformed into the intermediates of the TCA cycle.
- F1 pathway: P.putida is able to undertake this pathway, which consists in the introduction of two hydroxyl groups into toluene, forming cis-toluene dihydrodiol. This intermediate is then converted to 3-methylcatechol.
- KR1 pathway: Pseudomonas mendocina KR1, is able to convert toluene into p-cresol, by the enzyme toluene 4-monooxygenase. Subsequently, p-hydroxybenzoate is formed through oxidation of the methyl side chain.
- PK01: Pseudomonas pickettii PKO1 oxidizes toluene with the enzyme toluene 3-monooxygease to m-cresol, which is further oxidized to 3-methylcatechol by another monooxygenase.
- G4: The G4 pathway was observed in Bukholderia cepacia G4, where toluene is converted into o-cresol by toluene 2-monooxygenase and subsequently another monooxygenase converts it to 3-methylcatechol.
Anaerobic degradation pathways
Oil components that are trapped in marine sediments tend to persist in anaerobic conditions. Some hydrocarbons can be oxidized under anaerobic conditions when nitrate reduction, sulfate reduction, methane production, Fe3+ reduction or photosynthesis are coupled to hydrocarbon oxidation.[16]
Anaerobic bacterium strain HD-1 grows on CO2 in the presence of H
2 or tetradecane. In the absence of H
2, tetradecane is degraded, and the major metabolic intermediate is 1-dodecene[17]

Factors influencing the biodegradation
The biodegradation of hydrocarbons is limited by a number of chemical, physical and biological factors.

- Biosurfactants:[19] The contact between bacteria and hydrocarbons is fundamental because the first degradative step involves the use of oxygenase. Contact is favored by adhesion and emulsifying mechanisms[20] Bacteria that break down hydrocarbons often produce bioemulsifiers as secondary metabolites. These can be divided into low molecular weight molecules which effectively lower surface and interfacial tensions, and high molecular weight polymers which bond firmly to surfaces. Some bioemulsifiers promote the growth of bacteria on hydrophobic substrates insoluble in water by increasing their bioavailability, increasing their surface, desorbing them from surfaces and increasing their solubility. Biosurfactants have an amphiphilic nature, which allows the microorganisms that produce them to exploit hydrophobic substrates, allowing motility, avoiding competitors. The hydrophobic part usually comprises saturated or unsaturated fatty acids, fatty hydroxy acids or fatty alcohols with a chain length between 8 and 18 carbon atoms. The hydrophilic components consist either of small hydroxyl, phosphate or carboxylic groups, or of portions of carbohydrates or peptides. Biosurfactants are predominantly anionic and non-ionic compounds.
- Nutritional requirements for growth: The HCB also need large amounts of nitrogen and phosphorus. It has been estimated that 150 g of nitrogen and 30 g of phosphorus are required for 1 kg of oxidized hydrocarbon.[5]
- Temperature: The biodegradation of petroleum hydrocarbons occurs in an optimum temperature between 20 °C and 50 °C, while at lower temperatures the deterioration rate is slower.[21]
- pH and oxygen: Bacteria require a neutral pH, and in this the same oil can help neutralize environments that are too acidic for microbial growth. Oxygen is critical for aerobic degradation.[22]
Ecology
At the moment, most studies of the ecology of hydrocarbonoclastic bacteria refer to a wide group of genera found principally in marine environments. Since each of them is characterized by a different metabolism, these organisms work together in order to degrade all types of hydrocarbon compounds in a very efficient way. They also play a fundamental role in the carbon biogeochemical cycle and several studies show that some species can create intricate relationships with different marine organisms.[23]
Oil degrading microbial communities and ecological successions
When a release of oil (or whichever kind of hydrocarbon compound) happens in a specific marine area, a lot of bacterial species begin to colonize it, changing the microbial community already present there. Analyzing the dynamics of those communities has led to the discovery of common patterns that are associated with biodegradation, and those information can be useful for the improving of bioremediation methods. Microbial community in situ shuffles since the quantities of nutrients change as the presence of hydrocarbons increases: this ecological situation is able to select only those organisms which can use hydrocarbons as an energy source and possess all the enzymes to do so. In addition, most oil biodegrading species require specific quantities of phosphorus and nitrogen to carry out their catabolic processes. It is possible to state therefore, that hydrocarbonoclastic bacteria rate is limited by the availability of nitrogen and phosphorus mainly.[24]
Several experiments conducted both in vitro and in situ showed the fundamental role that OHCBs (obligate hydrocarbonoclastic bacteria) play during events like an oil spill. The very first microorganism populations that bloom when hydrocarbons are released are the so-called generalists, which can break (through specific enzymes) the most simple bonds in hydrocarbons (generally they are n-alkane degraders); among them, the most common genus is Alcanivorax (the most important species is Alcanivorax borkumensis) which can degrade aliphatic hydrocarbon compounds.[25] Subsequently, specialists replace generalists to degrade stronger and more complex bounds; among them one of the most known genera is Cycloclasticus which can, for example, degrade aromatic hydrocarbons such as PAH (polycyclic aromatic hydrocarbons).[26]
Up to now, no hydrocarbonoclastic Archaea species have been found, since it appears that they are too sensitive to the effects of an oil spill, as shown by many studies carried out on beaches and coastal waters.[27] Nevertheless, Archaea species could be used as markers of the ecological status of an environment.
Relationships with other marine organisms during bioremediation processes
Hydrocarbonoclastic bacteria form just a part of the ecological network during bioremediation and biodegradation processes, which involves many direct and indirect relationships and interactions with other communities and with the surrounding environment too. Such interactions include competition for limiting nutrients, predation by protozoa, lysis by phages and cooperative interactions that can decrease or increase degradation of hydrocarbons.[23]
Nutrients availability, as well as nutrients recycling, are important aspects of biodegradation communities. As said before, the amount of phosphorus and nitrogen can modify the structure of a microbial populations and consequently the composition of the community that is shaped by the presence of certain molecules in the ecosystem.
Predation and interactions with phages also affect development of a hydrocarbonoclastic bacterial community. It is possible that the increase of the turnover of biomass (which can be obtained by stimulating the activity of bacteriophages lysis or protozoa predation) could benefit hydrocarbonoclastic populations by stimulating biological remediation.[28] In fact, the presence of oil in the environment can induce prophages[29] and the subsequent lysis of a huge number of bacteria.[30] At the same time, nutrients recycling caused by phages' lysis can trigger a bloom of those species who are favored by the presence of both nutrients and hydrocarbons (used as energy resource). On the other hand, the presence of protozoa can create the opposite situation (it has a negative effect on biodegradation), by limiting the growth of bacterial populations in the ecosystem.[31] That is why interactions with predators are fundamental in marine environments. Nevertheless, in specific occasions, the presence of predators can boost bacterial degradation, as it happens for benzene[32] or toluene.[33] Moreover, in a similar way to what happens with phages, the activity of predation does create a nutritional loop, because predators can remineralize nutrients, which increases bacterial growth.
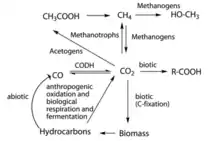
Role in biogeochemical carbon cycle
Since hydrocarbonoclastic bacteria can oxidize long carbon compounds, their metabolism includes part of the large family of biotic reactions in the biogeochemical carbon cycle. Hydrocarbons, especially alkanes, are produced by myriad organisms as waste, for defense, as structural elements, and as chemoattractants.[34] Therefore, this type of biodegradation represents one of the major sinks of hydrocarbon compounds and one of the source of carbon dioxide in marine environments.
In conclusion, hydrocarbonoclastic bacteria can mobilize hydrocarbons from natural sources and use the oxidized carbon atoms and introduce them into their metabolic central pathways. Those oxidized molecules enter the biotic phase of the carbon cycle and can be assimilated by primary and secondary consumers through predation or can assume them after cells' death.
Biotechnological applications
Hydrocarbon-degrading bacteria have many different applications but has specially importance their role in the field of environmental microbiology.[35]
Marine hydrocarbonoclastic bacteria are powerful tools for bioremediation, as they can degrade and convert contaminant oils because of their catabolic versatility.[36] In that way, using biotechnology is possible accelerate the cleaning up of a contaminated site such as coastal regions and offshore after an oil spills or human activities' pollution, but also it is possible to contain and mitigate their damage.[37] They normally bloom after an oil spill or other pollution, and because as they are very versatile metabolically, they can grow on minimal mediums. One example of this is the nitrogen-fixing and heavy oil-degrading bacterium Azospirillum oleiclasticum, which was isolated from an oil production mixture.[38] But A. oleiclasticum is not the only strain that can grow on oil, a 2013 study discovered that there are at least 125 strains, adapted to grow on minimal medium supplemented with crude oil. The predominant bacterial detected by approaches were the Proteobacteria and the most abundant species were in genera Acinetobacter and Stenotrophomons.[39]
They are also used in biosynthesis because they are an extraordinary archive of enzymes like mono and dioxygenases, oxidases, dehydrogenases and others. Furthermore, as they are adapted to grow in hydrocarbon-rich environments, they often synthesize characteristic compounds like polymeric storage substances of industrial interest and bio-detergents with high emulsifying activity. One example of this is the use of the oleaginous yeast Yarrowia lipolytica. As this yeast has a versatile lipid metabolism, by its combination with specific bacterial genes it can use specific enzymatic pathways to bioconvert different lipids (petroleum, alkane, vegetable oil, fatty acid), fats and oils into industrially valuable lipid-derived compounds like isoprenoid-derived compounds (carotenoids, polyenic carotenoid ester), wax esters (WE), polyhydroxyalkanoates (PHAs) and free hydroxylated fatty acids (HFAs).[40]
References
- 1 2 Das, Nilanjana; Chandran, Preethy (2011-09-13). "Microbial Degradation of Petroleum Hydrocarbon Contaminants: An Overview". Biotechnology Research International. 2011: 941810. doi:10.4061/2011/941810. ISSN 2090-3138. PMC 3042690. PMID 21350672.
- ↑ Bacosa, Hernando P.; Kamalanathan, Manoj; Chiu, Meng-Hsuen; Tsai, Shih-Ming; Sun, Luni; Labonté, Jessica M.; Schwehr, Kathleen A.; Hala, David; Santschi, Peter H.; Chin, Wei-Chun; Quigg, Antonietta (2018-12-06). "Extracellular polymeric substances (EPS) producing and oil degrading bacteria isolated from the northern Gulf of Mexico". PLOS ONE. 13 (12): e0208406. Bibcode:2018PLoSO..1308406B. doi:10.1371/journal.pone.0208406. ISSN 1932-6203. PMC 6283562. PMID 30521589.
- ↑ Head, Ian M.; Jones, D. Martin; Röling, Wilfred F. M. (March 2006). "Marine microorganisms make a meal of oil". Nature Reviews Microbiology. 4 (3): 173–182. doi:10.1038/nrmicro1348. ISSN 1740-1526. PMID 16489346. S2CID 251141.
- ↑ Gibson, D. T.; Parales, R. E. (June 2000). "Aromatic hydrocarbon dioxygenases in environmental biotechnology". Current Opinion in Biotechnology. 11 (3): 236–243. doi:10.1016/s0958-1669(00)00090-2. ISSN 0958-1669. PMID 10851146.
- 1 2 Rosenberg, E.; Navon-Venezia, S.; Zilber-Rosenberg, I.; Ron, E. Z. (1998). "Rate-Limiting Steps in the Microbial Degradation of Petroleum Hydrocarbons". Soil and Aquifer Pollution. pp. 159–172. doi:10.1007/978-3-662-03674-7_11. ISBN 978-3-642-08294-8.
- ↑ Maruyama, A.; Ishiwata, H.; Kitamura, K.; Sunamura, M.; Fujita, T.; Matsuo, M.; Higashihara, T. (2003). "Dynamics of microbial populations and strong selection for Cycloclasticus pugetii following the Nakhodka oil spill". Microbial Ecology. 46 (4): 442–453. doi:10.1007/s00248-002-3010-z. ISSN 0095-3628. PMID 12904913. S2CID 21849775.
- ↑ Ron, Eliora Z.; Rosenberg, Eugene (June 2014). "Enhanced bioremediation of oil spills in the sea". Current Opinion in Biotechnology. 27: 191–194. doi:10.1016/j.copbio.2014.02.004. ISSN 1879-0429. PMID 24657912.
- ↑ J. J. Perry, “Microbial metabolism of cyclic alkanes,” in Petroleum Microbiology, R. M. Atlas, Ed., pp. 61–98, Macmil- lan, New York, NY, USA, 1984.
- ↑ W. Ulrici, “Contaminant soil areas, different countries and contaminant monitoring of contaminants,” in Environmental Process II. Soil Decontamination Biotechnology, H. J. Rehm and G. Reed, Eds., vol. 11, pp. 5–42, 2000.
- 1 2 3 Harayama, S.; Kishira, H.; Kasai, Y.; Shutsubo, K. (1999). "Petroleum biodegradation in marine environments". Journal of Molecular Microbiology and Biotechnology. 1 (1): 63–70. ISSN 1464-1801. PMID 10941786.
- ↑ Van Beilen, Jan B.; Wubbolts, Marcel G.; Witholt, Bernard (1994). "Genetics of alkane oxidation by Pseudomonas oleovorans" (PDF). Biodegradation. 5 (3–4): 161–174. doi:10.1007/BF00696457. PMID 7532480. S2CID 30001069.
- ↑ Markovetz, A. J.; Kallio, Reino E. (1971). "Subterminal Oxidation of Aliphatic Hydrocarbons by Microorganisms". CRC Critical Reviews in Microbiology. 1 (2): 225–237. doi:10.3109/10408417109104482.
- ↑ Morgan, Philip; Watkinson, Robert J. (1994). "Biodegradation of components of petroleum". Biochemistry of microbial degradation. pp. 1–31. doi:10.1007/978-94-011-1687-9_1. ISBN 978-94-010-4738-8.
- ↑ Cafaro, Valeria; Izzo, Viviana; Notomista, Eugenio; Di Donato, Alberto (2013-01-01), Trincone, Antonio (ed.), "14 - Marine hydrocarbonoclastic bacteria", Marine Enzymes for Biocatalysis, Woodhead Publishing Series in Biomedicine, Woodhead Publishing, pp. 373–402, ISBN 978-1-907568-80-0, retrieved 2022-07-19
- ↑ Johnson, G. R.; Olsen, R. H. (1997). "Multiple pathways for toluene degradation in Burkholderia sp. Strain JS150". Applied and Environmental Microbiology. 63 (10): 4047–4052. Bibcode:1997ApEnM..63.4047J. doi:10.1128/aem.63.10.4047-4052.1997. PMC 168715. PMID 9327568.
- ↑ Evans, W. C.; Fuchs, G. (1988). "Anaerobic Degradation of Aromatic Compounds". Annual Review of Microbiology. 42: 289–317. doi:10.1146/annurev.mi.42.100188.001445. PMID 3059996.
- ↑ Morikawa, Masaaki; Kanemoto, Mitsuhide; Imanaka, Tadayuki (1996-01-01). "Biological oxidation of alkane to alkene under anaerobic conditions". Journal of Fermentation and Bioengineering. 82 (3): 309–311. doi:10.1016/0922-338X(96)88825-8. ISSN 0922-338X.
- ↑ Das, Nilanjana; Chandran, Preethy (2011). "Microbial Degradation of Petroleum Hydrocarbon Contaminants: An Overview". Biotechnology Research International. 2011: 941810. doi:10.4061/2011/941810. PMC 3042690. PMID 21350672.
- ↑ Kubicki, Sonja; Bollinger, Alexander; Katzke, Nadine; Jaeger, Karl-Erich; Loeschcke, Anita; Thies, Stephan (2019). "Marine Biosurfactants: Biosynthesis, Structural Diversity and Biotechnological Applications". Marine Drugs. 17 (7): 408. doi:10.3390/md17070408. ISSN 1660-3397. PMC 6669457. PMID 31323998.
- ↑ Rosenberg, E. (1992). "The hydrocarbon-oxidizing bacteria". In Rosenberg, E.; DeLong, E.; Thompson, F.; Lorey, S.; Stackebrandt, E. (eds.). The Prokaryotes: A Handbook on the Biology of Bacteria: Ecophysiology, Isolation, Identification, Applications (4th ed.). New York: Springer-Verlag Inc. (published 2012). ISBN 9780387972589.
- ↑ Bartha, R. and Bossert, I. (1984). The treatment and disposal of petroleum wastes. In: Petroleum Microbiology, R. M. Atlas, Macmillan, New York, NY, USA. 553–578.
- ↑ Van Hamme, Jonathan D.; Singh, Ajay; Ward, Owen P. (2003). "Recent Advances in Petroleum Microbiology". Microbiology and Molecular Biology Reviews. 67 (4): 503–549. doi:10.1128/MMBR.67.4.503-549.2003. PMC 309048. PMID 14665675.
- 1 2 Head, Ian M.; Jones, D. Martin; Röling, Wilfred F. M. (March 2006). "Marine microorganisms make a meal of oil". Nature Reviews Microbiology. 4 (3): 173–182. doi:10.1038/nrmicro1348. ISSN 1740-1534. PMID 16489346. S2CID 251141.
- ↑ Atlas, R. M.; Bartha, R. (May 1972). "Degradation and mineralization of petroleum in sea water: limitation by nitrogen and phosphorous". Biotechnology and Bioengineering. 14 (3): 309–318. doi:10.1002/bit.260140304. ISSN 0006-3592. PMID 5029877. S2CID 23279895.
- ↑ Kasai, Yuki; Kishira, Hideo; Sasaki, Tetsuya; Syutsubo, Kazuaki; Watanabe, Kazuya; Harayama, Shigeaki (2002-03-05). "Predominant growth of Alcanivorax strains in oil-contaminated and nutrient-supplemented sea water". Environmental Microbiology. 4 (3): 141–147. doi:10.1046/j.1462-2920.2002.00275.x. ISSN 1462-2912. PMID 12000314.
- ↑ Kasai, Yuki; Kishira, Hideo; Harayama, Shigeaki (November 2002). "Bacteria belonging to the genus cycloclasticus play a primary role in the degradation of aromatic hydrocarbons released in a marine environment". Applied and Environmental Microbiology. 68 (11): 5625–5633. Bibcode:2002ApEnM..68.5625K. doi:10.1128/AEM.68.11.5625-5633.2002. ISSN 0099-2240. PMC 129893. PMID 12406758.
- ↑ Röling, Wilfred F. M.; Couto de Brito, Ivana R.; Swannell, Richard P. J.; Head, Ian M. (May 2004). "Response of Archaeal Communities in Beach Sediments to Spilled Oil and Bioremediation". Applied and Environmental Microbiology. 70 (5): 2614–2620. Bibcode:2004ApEnM..70.2614R. doi:10.1128/AEM.70.5.2614-2620.2004. ISSN 0099-2240. PMC 404411. PMID 15128510.
- ↑ Röling, Wilfred F. M.; Milner, Michael G.; Jones, D. Martin; Lee, Kenneth; Daniel, Fabien; Swannell, Richard J. P.; Head, Ian M. (2002). "Robust hydrocarbon degradation and dynamics of bacterial communities during nutrient-enhanced oil spill bioremediation". Applied and Environmental Microbiology. 68 (11): 5537–5548. Bibcode:2002ApEnM..68.5537R. doi:10.1128/AEM.68.11.5537-5548.2002. ISSN 0099-2240. PMC 129918. PMID 12406747.
- ↑ Cochran, P. K.; Kellogg, C. A.; Paul, J. H. (1998-10-23). "Prophage induction of indigenous marine lysogenic bacteria by environmental pollutants". Marine Ecology Progress Series. 164: 125–133. Bibcode:1998MEPS..164..125C. doi:10.3354/meps164125.
- ↑ Jiang, SC; Paul, JH (1996). "Occurrence of lysogenic bacteria in marine microbial communities as determined by prophage induction". Marine Ecology Progress Series. 142: 27–38. Bibcode:1996MEPS..142...27J. doi:10.3354/meps142027. ISSN 0171-8630.
- ↑ Kota, Sreenivas; Borden, Robert C; Barlaz, Morton A (1999-06-01). "Influence of protozoan grazing on contaminant biodegradation". FEMS Microbiology Ecology. 29 (2): 179–189. doi:10.1111/j.1574-6941.1999.tb00609.x. ISSN 0168-6496.
- ↑ Mattison, R. G.; Taki, H.; Harayama, S. (January 2005). "The soil flagellate Heteromita globosa accelerates bacterial degradation of alkylbenzenes through grazing and acetate excretion in batch culture". Microbial Ecology. 49 (1): 142–150. doi:10.1007/s00248-003-0226-5. ISSN 0095-3628. PMID 15690226. S2CID 8180381.
- ↑ Mattison, R. G.; Harayama, S. (2001-01-01). "The predatory soil flagellate Heteromita globosa stimulates toluene biodegradation by a Pseudomonas sp". FEMS Microbiology Letters. 194 (1): 39–45. doi:10.1111/j.1574-6968.2001.tb09443.x. ISSN 0378-1097. PMID 11150663. S2CID 21268831.
- ↑ Austin, Rachel Narehood; Groves, John T (2011-08-01). "Alkane-oxidizing metalloenzymes in the carbon cycle". Metallomics. 3 (8): 775–787. doi:10.1039/c1mt00048a. ISSN 1756-5901. PMID 21743926.
- ↑ "Marine Enzymes for Biocatalysis. Sources, Biocatalytic Characteristics and Bioprocesses of Marine Enzymes". Woodhead Publishing Series in Biomedicine, 2013: 373–402.
- ↑ Anal Chavan, Suparna Mukherji (2008). "Treatment of hydrocarbon-rich wastewater using oil degrading bacteria and phototrophic microorganisms in rotating biological contactor: Effect of N:P ratio". Journal of Hazardous Materials. 154 (1–3): 63–72. doi:10.1016/j.jhazmat.2007.09.106. PMID 17983704.
- ↑ Sonja Kubicki, Alexander Bollinger, Nadine Katzke, Karl-Erich Jaeger, Anita Loeschcke, and Stephan Thies (2019). "Marine Biosurfactants: Biosynthesis, Structural Diversity and Biotechnological Applications". Marine Drugs. 17 (7): 408. doi:10.3390/md17070408. PMC 6669457. PMID 31323998.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Wu, Danni; Zhang, Xiao-Juan; Liu, Hong-Can; Zhou, Yu-Guang; Wu, Xiao-Lei; Nie, Yong; Kang, Ying-Qian; Cai, Man (2021). "Azospirillum oleiclasticum sp. nov, a nitrogen-fixing and heavy oil degrading bacterium isolated from an oil production mixture of Yumen Oilfield". Systematic and Applied Microbiology. 44 (1): 126171. doi:10.1016/j.syapm.2020.126171. PMID 33360414. S2CID 229448870.
- ↑ Mahjoubi, Mouna; Jaouani, Atef; Guesmi, Amel; Ben Amor, Sonia; Jouini, Ahlem; Cherif, Hanen; Najjari, Afef; Boudabous, Abdellatif; Koubaa, Nedra; Cherif, Ameur (September 2013). "Hydrocarbonoclastic bacteria isolated from petroleum contaminated sites in Tunisia: isolation, identification and characterization of the biotechnological potential". New Biotechnology. 30 (6): 723–733. doi:10.1016/j.nbt.2013.03.004. PMID 23541698.
- ↑ Julia S. Sabirova, R. Haddouche, I. N. Van Bogaert, F. Mulaa, W. Verstraete, K. N. Timmis, C. Schmidt-Dannert, J. M. Nicaud, and W. Soetaert (2011). "The 'LipoYeasts' project: using the oleaginous yeast Yarrowia lipolytica in combination with specific bacterial genes for the bioconversion of lipids, fats and oils into high-value products". Microbial Biotechnology. 4 (1): 47–54. doi:10.1111/j.1751-7915.2010.00187.x. PMC 3815794. PMID 21255371.
{{cite journal}}: CS1 maint: multiple names: authors list (link)
External links
 Media related to Hydrocarbonoclastic bacteria at Wikimedia Commons
Media related to Hydrocarbonoclastic bacteria at Wikimedia Commons