| CRISPR-associated protein 4 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 | |||||||||
| Identifiers | |||||||||
| Symbol | cas4 | ||||||||
| Pfam | PF01930 | ||||||||
| InterPro | IPR022765 | ||||||||
| |||||||||
Cas4 is an endonuclease found in some CRISPR systems that is used as part of the spacer acquisition stage.
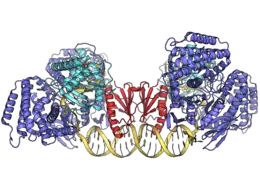
Geobacter sulfurreducens Cas1-Cas2-Cas4 complex bound to double stranded protospacer adjacent motif (PAM) spacer DNA. Cas1 homodimers (blue) bind to each side of the Cas2 homodimer (red) with a Cas4 Fe-S nuclease (cyan) bridging the two. The 3′ end of each DNA strand threads into the Cas4 active site. PDB: 7MI4
Cas4 is required for efficient prespacer processing by forming a Cas4-Cas1-Cas2 complex. [1] In this complex, Cas4 cuts double-stranded substrates with long 3'-overhangs through site-specific endonucleolytic cleavage. [2] Cas4 closely interacts with the integrase active sites of Cas1, recognizes PAM sequences within the substrate and cleaves precisely upstream of them, ensuring the acquisition of functional spacers. [2] [1]
References
- 1 2 Lee H, Dhingra Y, Sashital DG (April 2019). "The Cas4-Cas1-Cas2 complex mediates precise prespacer processing during CRISPR adaptation". eLife. 8. doi:10.7554/eLife.44248. PMC 6519985. PMID 31021314.
- 1 2 Lee H, Zhou Y, Taylor DW, Sashital DG (April 2018). "Cas4-Dependent Prespacer Processing Ensures High-Fidelity Programming of CRISPR Arrays". Molecular Cell. 70 (1): 48–59.e5. doi:10.1016/j.molcel.2018.03.003. PMC 5889325. PMID 29602742.
This article is issued from Wikipedia. The text is licensed under Creative Commons - Attribution - Sharealike. Additional terms may apply for the media files.