| Part of a series on |
| Genetic engineering |
|---|
 |
| Genetically modified organisms |
| History and regulation |
| Process |
| Applications |
| Controversies |
Genetically modified bacteria were the first organisms to be modified in the laboratory, due to their simple genetics.[1] These organisms are now used for several purposes, and are particularly important in producing large amounts of pure human proteins for use in medicine.[2]
History
The first example of this occurred in 1978 when Herbert Boyer, working at a University of California laboratory, took a version of the human insulin gene and inserted into the bacterium Escherichia coli to produce synthetic "human" insulin. Four years later, it was approved by the U.S. Food and Drug Administration.
Research

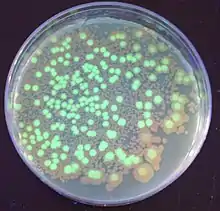
Bacteria were the first organisms to be genetically modified in the laboratory, due to the relative ease of modifying their chromosomes.[3] This ease made them important tools for the creation of other GMOs. Genes and other genetic information from a wide range of organisms can be added to a plasmid and inserted into bacteria for storage and modification. Bacteria are cheap, easy to grow, clonal, multiply quickly, are relatively easy to transform, and can be stored at -80 °C almost indefinitely. Once a gene is isolated it can be stored inside the bacteria, providing an unlimited supply for research.[4] The large number of custom plasmids make manipulating DNA excised from bacteria relatively easy.[5]
Their ease of use has made them great tools for scientists looking to study gene function and evolution. Most DNA manipulation takes place within bacterial plasmids before being transferred to another host. Bacteria are the simplest model organism and most of our early understanding of molecular biology comes from studying Escherichia coli.[6] Scientists can easily manipulate and combine genes within the bacteria to create novel or disrupted proteins and observe the effect this has on various molecular systems. Researchers have combined the genes from bacteria and archaea, leading to insights on how these two diverged in the past.[7] In the field of synthetic biology, they have been used to test various synthetic approaches, from synthesizing genomes to creating novel nucleotides.[8][9][10]
Food
Bacteria have been used in the production of food for a very long time, and specific strains have been developed and selected for that work on an industrial scale. They can be used to produce enzymes, amino acids, flavourings, and other compounds used in food production. With the advent of genetic engineering, new genetic changes can easily be introduced into these bacteria. Most food-producing bacteria are lactic acid bacteria, and this is where the majority of research into genetically engineering food-producing bacteria has gone. The bacteria can be modified to operate more efficiently, reduce toxic byproduct production, increase output, create improved compounds, and remove unnecessary pathways.[11] Food products from genetically modified bacteria include alpha-amylase, which converts starch to simple sugars, chymosin, which clots milk protein for cheese making, and pectinesterase, which improves fruit juice clarity.[12]
In cheese
Chymosin is an enzyme produced in the stomach of young ruminant mammals to digest milk. The digestion of milk proteins via enzymes is essential to cheesemaking. The species Escherichia coli and Bacillus subtilis can be genetically engineered to synthesise and excrete chymosin,[13] providing a more efficient means of production. The use of bacteria to synthesise chymosin also provides a vegetarian method of cheesemaking, as previously, young ruminants (typically calves) had to be slaughtered to extract the enzyme from the stomach lining.
Industrial
Genetically modified bacteria are used to produce large amounts of proteins for industrial use. Generally the bacteria are grown to a large volume before the gene encoding the protein is activated. The bacteria are then harvested and the desired protein purified from them.[14] The high cost of extraction and purification has meant that only high value products have been produced at an industrial scale.[15]
Pharmaceutical production
The majority of the industrial products from bacteria are human proteins for use in medicine.[16] Many of these proteins are impossible or difficult to obtain via natural methods and they are less likely to be contaminated with pathogens, making them safer.[14] Prior to recombinant protein products, several treatments were derived from cadavers or other donated body fluids and could transmit diseases.[17] Indeed, transfusion of blood products had previously led to unintentional infection of haemophiliacs with HIV or hepatitis C; similarly, treatment with human growth hormone derived from cadaver pituitary glands may have led to outbreaks of Creutzfeldt–Jakob disease.[17][18]
The first medicinal use of GM bacteria was to produce the protein insulin to treat diabetes.[19] Other medicines produced include clotting factors to treat haemophilia,[20] human growth hormone to treat various forms of dwarfism,[21][22] interferon to treat some cancers, erythropoietin for anemic patients, and tissue plasminogen activator which dissolves blood clots.[14] Outside of medicine they have been used to produce biofuels.[23] There is interest in developing an extracellular expression system within the bacteria to reduce costs and make the production of more products economical.[15]
Health
With greater understanding of the role that the microbiome plays in human health, there is the potential to treat diseases by genetically altering the bacteria to, themselves, be therapeutic agents. Ideas include altering gut bacteria so they destroy harmful bacteria, or using bacteria to replace or increase deficient enzymes or proteins. One research focus is to modify Lactobacillus, bacteria that naturally provide some protection against HIV, with genes that will further enhance this protection.[24] The bacteria which generally cause tooth decay have been engineered to no longer produce tooth-corroding lactic acid.[25] These transgenic bacteria, if allowed to colonize a person's mouth, could perhaps reduce the formation of cavities.[26] Transgenic microbes have also been used in recent research to kill or hinder tumors, and to fight Crohn's disease.[27]
If the bacteria do not form colonies inside the patient, the person must repeatedly ingest the modified bacteria in order to get the required doses. Enabling the bacteria to form a colony could provide a more long-term solution, but could also raise safety concerns as interactions between bacteria and the human body are less well understood than with traditional drugs.
One example of such an intermediate, which only forms short-term colonies in the gastrointestinal tract, may be Lactobacillus Acidophilus MPH734. This is used as a specific in the treatment of Lactose Intolerance. This genetically modified version of Lactobacillus acidophilus bacteria produces a missing enzyme called lactase which is used for the digestion of lactose found in dairy products or, more commonly, in food prepared with dairy products. The short term colony is induced over a one-week, 21-pill treatment regimen, after which, the temporary colony can produce lactase for three months or more before it is removed from the body by a natural processes. The induction regimen can be repeated as often as necessary to maintain protection from the symptoms of lactose intolerance, or discontinued with no consequences, except the return of the original symptoms.
There are concerns that horizontal gene transfer to other bacteria could have unknown effects. As of 2018 there are clinical trials underway testing the efficacy and safety of these treatments.[24]
Agriculture
For over a century bacteria have been used in agriculture. Crops have been inoculated with Rhizobia (and more recently Azospirillum) to increase their production or to allow them to be grown outside their original habitat. Application of Bacillus thuringiensis (Bt) and other bacteria can help protect crops from insect infestation and plant diseases. With advances in genetic engineering, these bacteria have been manipulated for increased efficiency and expanded host range. Markers have also been added to aid in tracing the spread of the bacteria. The bacteria that naturally colonise certain crops have also been modified, in some cases to express the Bt genes responsible for pest resistance. Pseudomonas strains of bacteria cause frost damage by nucleating water into ice crystals around themselves. This led to the development of ice-minus bacteria, that have the ice-forming genes removed. When applied to crops they can compete with the ice-plus bacteria and confer some frost resistance.[28]

Other uses
Other uses for genetically modified bacteria include bioremediation, where the bacteria are used to convert pollutants into a less toxic form. Genetic engineering can increase the levels of the enzymes used to degrade a toxin or to make the bacteria more stable under environmental conditions.[29] GM bacteria have also been developed to leach copper from ore,[30] clean up mercury pollution[31] and detect arsenic in drinking water.[32] Bioart has also been created using genetically modified bacteria. In the 1980s artist Joe Davis and geneticist Dana Boyd converted the Germanic symbol for femininity (ᛉ) into binary code and then into a DNA sequence, which was then expressed in Escherichia coli.[33] This was taken a step further in 2012, when a whole book was encoded onto DNA.[34] Paintings have also been produced using bacteria transformed with fluorescent proteins.[33][35][36]
Bacteria-synthesized transgenic products
References
- ↑ Melo EO, Canavessi AM, Franco MM, Rumpf R (2007). "Animal transgenesis: state of the art and applications" (PDF). Journal of Applied Genetics. 48 (1): 47–61. doi:10.1007/BF03194657. PMID 17272861. S2CID 24578435. Archived from the original (PDF) on 6 November 2009.
- ↑ Leader B, Baca QJ, Golan DE (January 2008). "Protein therapeutics: a summary and pharmacological classification". Nature Reviews. Drug Discovery. A guide to drug discovery. 7 (1): 21–39. doi:10.1038/nrd2399. PMID 18097458. S2CID 3358528.
- ↑ Melo EO, Canavessi AM, Franco MM, Rumpf R (2007). "Animal transgenesis: state of the art and applications" (PDF). Journal of Applied Genetics. 48 (1): 47–61. doi:10.1007/BF03194657. PMID 17272861. S2CID 24578435. Archived from the original (PDF) on 6 November 2009.
- ↑ "Rediscovering Biology - Online Textbook: Unit 13 Genetically Modified Organisms". www.learner.org. Archived from the original on 2019-12-03. Retrieved 2017-08-18.
- ↑ Fan M, Tsai J, Chen B, Fan K, LaBaer J (March 2005). "A central repository for published plasmids". Science. 307 (5717): 1877. doi:10.1126/science.307.5717.1877a. PMID 15790830. S2CID 27404861.
- ↑ Cooper GM (2000). "Cells As Experimental Models". The Cell: A Molecular Approach. 2nd Edition.
- ↑ Patel P (June 2018). "Microbe Mystery". Scientific American. 319 (1): 18. Bibcode:2018SciAm.319a..18P. doi:10.1038/scientificamerican0718-18a. PMID 29924081. S2CID 49310760.
- ↑ Arpino JA, Hancock EJ, Anderson J, Barahona M, Stan GB, Papachristodoulou A, Polizzi K (July 2013). "Tuning the dials of Synthetic Biology". Microbiology. 159 (Pt 7): 1236–53. doi:10.1099/mic.0.067975-0. PMC 3749727. PMID 23704788.
- ↑ Pollack A (7 May 2014). "Researchers Report Breakthrough in Creating Artificial Genetic Code". The New York Times. Retrieved 7 May 2014.
- ↑ Malyshev DA, Dhami K, Lavergne T, Chen T, Dai N, Foster JM, Corrêa IR, Romesberg FE (May 2014). "A semi-synthetic organism with an expanded genetic alphabet". Nature. 509 (7500): 385–8. Bibcode:2014Natur.509..385M. doi:10.1038/nature13314. PMC 4058825. PMID 24805238.
- ↑ Kärenlampi SO, von Wright AJ (2016-01-01). Encyclopedia of Food and Health. pp. 211–216. doi:10.1016/B978-0-12-384947-2.00356-1. ISBN 9780123849533.
- ↑ Panesar, Pamit et al. (2010) Enzymes in Food Processing: Fundamentals and Potential Applications, Chapter 10, I K International Publishing House, ISBN 978-93-80026-33-6
- ↑ Luerce, T.D.; Azevedo, M. S.; LeBlanc, J.G.; Azevedo, V.; Miyoshi, A.; Pontes, D. S. (November–December 2014). "Recombinant Lactococcus lactis fails to secrete bovine chymosine". Bioengineered. 5 (6): 363–370. doi:10.4161/bioe.36327. PMC 4601287. PMID 25482140.
- 1 2 3 Jumba M (2009). Genetically Modified Organisms the Mystery Unraveled. Durham: Eloquent Books. pp. 51–54. ISBN 9781609110819.
- 1 2 Zhou Y, Lu Z, Wang X, Selvaraj JN, Zhang G (February 2018). "Genetic engineering modification and fermentation optimization for extracellular production of recombinant proteins using Escherichia coli". Applied Microbiology and Biotechnology. 102 (4): 1545–1556. doi:10.1007/s00253-017-8700-z. PMID 29270732. S2CID 2694760.
- ↑ Leader B, Baca QJ, Golan DE (January 2008). "Protein therapeutics: a summary and pharmacological classification". Nature Reviews. Drug Discovery. A guide to drug discovery. 7 (1): 21–39. doi:10.1038/nrd2399. PMID 18097458. S2CID 3358528.
- 1 2 Foster PR (October 2000). "Prions and blood products". Annals of Medicine. 32 (7): 501–13. doi:10.3109/07853890009002026. PMID 11087171. S2CID 9331069.
- ↑ Key NS, Negrier C (August 2007). "Coagulation factor concentrates: past, present, and future". Lancet. 370 (9585): 439–48. doi:10.1016/S0140-6736(07)61199-4. PMID 17679021. S2CID 26527486.
- ↑ Walsh G (April 2005). "Therapeutic insulins and their large-scale manufacture". Applied Microbiology and Biotechnology. 67 (2): 151–9. doi:10.1007/s00253-004-1809-x. PMID 15580495. S2CID 5986035.
- ↑ Pipe SW (May 2008). "Recombinant clotting factors". Thrombosis and Haemostasis. 99 (5): 840–50. doi:10.1160/TH07-10-0593. PMID 18449413. S2CID 2701961.
- ↑ Bryant J, Baxter L, Cave CB, Milne R (July 2007). Bryant J (ed.). "Recombinant growth hormone for idiopathic short stature in children and adolescents" (PDF). The Cochrane Database of Systematic Reviews (3): CD004440. doi:10.1002/14651858.CD004440.pub2. PMID 17636758.
- ↑ Baxter L, Bryant J, Cave CB, Milne R (January 2007). Bryant J (ed.). "Recombinant growth hormone for children and adolescents with Turner syndrome" (PDF). The Cochrane Database of Systematic Reviews (1): CD003887. doi:10.1002/14651858.CD003887.pub2. PMID 17253498.
- ↑ Summers, Rebecca (24 April 2013) "Bacteria churn out first ever petrol-like biofuel" New Scientist, Retrieved 27 April 2013
- 1 2 Reardon S (June 2018). "Genetically modified bacteria enlisted in fight against disease". Nature. 558 (7711): 497–498. Bibcode:2018Natur.558..497R. doi:10.1038/d41586-018-05476-4. PMID 29946090.
- ↑ Hillman JD (August 2002). "Genetically modified Streptococcus mutans for the prevention of dental caries". Antonie van Leeuwenhoek. 82 (1–4): 361–6. doi:10.1023/A:1020695902160. PMID 12369203. S2CID 11066428.
- ↑ Hillman JD, Mo J, McDonell E, Cvitkovitch D, Hillman CH (May 2007). "Modification of an effector strain for replacement therapy of dental caries to enable clinical safety trials". Journal of Applied Microbiology. 102 (5): 1209–19. doi:10.1111/j.1365-2672.2007.03316.x. PMID 17448156.
- ↑ Braat H, Rottiers P, Hommes DW, Huyghebaert N, Remaut E, Remon JP, van Deventer SJ, Neirynck S, Peppelenbosch MP, Steidler L (June 2006). "A phase I trial with transgenic bacteria expressing interleukin-10 in Crohn's disease". Clinical Gastroenterology and Hepatology. 4 (6): 754–9. doi:10.1016/j.cgh.2006.03.028. PMID 16716759.
- ↑ Amarger N (November 2002). "Genetically modified bacteria in agriculture". Biochimie. 84 (11): 1061–72. doi:10.1016/s0300-9084(02)00035-4. PMID 12595134.
- ↑ Sharma B, Dangi AK, Shukla P (March 2018). "Contemporary enzyme based technologies for bioremediation: A review". Journal of Environmental Management. 210: 10–22. doi:10.1016/j.jenvman.2017.12.075. PMID 29329004.
- ↑ Valda D, Dowling J (10 December 2010). "Making Microbes Better Miners". Business Chile Magazine. Archived from the original on 17 December 2010. Retrieved 21 March 2012.
- ↑ Ruiz ON, Alvarez D, Gonzalez-Ruiz G, Torres C (August 2011). "Characterization of mercury bioremediation by transgenic bacteria expressing metallothionein and polyphosphate kinase". BMC Biotechnology. 11: 82. doi:10.1186/1472-6750-11-82. PMC 3180271. PMID 21838857.
- ↑ Sanderson K (24 February 2012). "New Portable Kit Detects Arsenic In Wells". Chemical and Engineering News.
- 1 2 Yetisen AK, Davis J, Coskun AF, Church GM, Yun SH (December 2015). "Bioart". Trends in Biotechnology. 33 (12): 724–734. doi:10.1016/j.tibtech.2015.09.011. PMID 26617334.
- ↑ Agapakis C. "Communicating with Aliens through DNA". Scientific American Blog Network. Retrieved 2018-09-13.
- ↑ Majdi, Mohammad; Ashengroph, Morahem; Abdollahi, Mohammad Reza (February 2016). "Sesquiterpene lactone engineering in microbial and plant platforms: parthenolide and artemisinin as case studies". Applied Microbiology and Biotechnology. 100 (3): 1041–1059. doi:10.1007/s00253-015-7128-6. ISSN 0175-7598. PMID 26567019. S2CID 9683430.
- ↑ McBride, William D.; El-Osta, Hisham S. (April 2002). "Impacts of the Adoption of Genetically Engineered Crops on Farm Financial Performance" (PDF). Journal of Agricultural and Applied Economics. 34 (1): 175–191. doi:10.1017/s1074070800002224. ISSN 1074-0708. S2CID 54910535.
- ↑ Joly-Guillou, Marie-Laure; Kempf, Marie; Cavallo, Jean-Didier; Chomarat, Monique; Dubreuil, Luc; Maugein, Jeanne; Muller-Serieys, Claudette; Roussel-Delvallez, Micheline (2010-03-18). "Comparative in vitro activity of Meropenem, Imipenem and Piperacillin/tazobactam against 1071 clinical isolates using 2 different methods: a French multicentre study". BMC Infectious Diseases. 10 (1): 72. doi:10.1186/1471-2334-10-72. ISSN 1471-2334. PMC 2845586. PMID 20298555.
Further reading
- Karl Drlica (2004). Understanding DNA and Gene Cloning: A Guide for the Curious (4th ed.). ISBN 978-0-471-43416-0.